Introduction to R
Week 4: Grouping and tables
Louisa Smith
August 3 - August 7
Let's
summarize
our data
Last week
We learned...
- Make a new variable with
mutate()
Last week
We learned...
- Make a new variable with
mutate() - Select the variables you want in your dataset with
select()
Last week
We learned...
- Make a new variable with
mutate() - Select the variables you want in your dataset with
select() - Keep only the observations you want in your dataset with
filter()
Last week
We learned...
- Make a new variable with
mutate() - Select the variables you want in your dataset with
select() - Keep only the observations you want in your dataset with
filter() - We also looked at categorizing our data with factors and the
forcatspackage
Your code might start to look like this
nlsy2 <- mutate(nlsy, only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no"))nlsy3 <- select(nlsy2, id, contains("sleep"), only)only_kids <- filter(nlsy3, only == "yes")only_kids## # A tibble: 30 x 4## id sleep_wkdy sleep_wknd only ## <dbl> <dbl> <dbl> <chr>## 1 458 7 8 yes ## 2 653 6 7 yes ## 3 1101 7 8 yes ## 4 1166 5 6 yes ## # … with 26 more rowsRepertoire of functions
We are doing more and more things to our dataset.
In any data management and/or analysis task, we perform a series of functions to the data until we get some object we want.
Sometimes this can be hard to read/keep track of.

Before we add another set of functions...
The pipe
Certain packages, including tidyverse, include a function known as a pipe.
If you have experience with unix programming, you may be familiar with the version of the pipe there: |.
R uses this as a pipe: %>%

The pipe function is originally from the magrittr package, named after René Magritte
We use the pipe to chain together steps
It's like a recipe for our dataset.
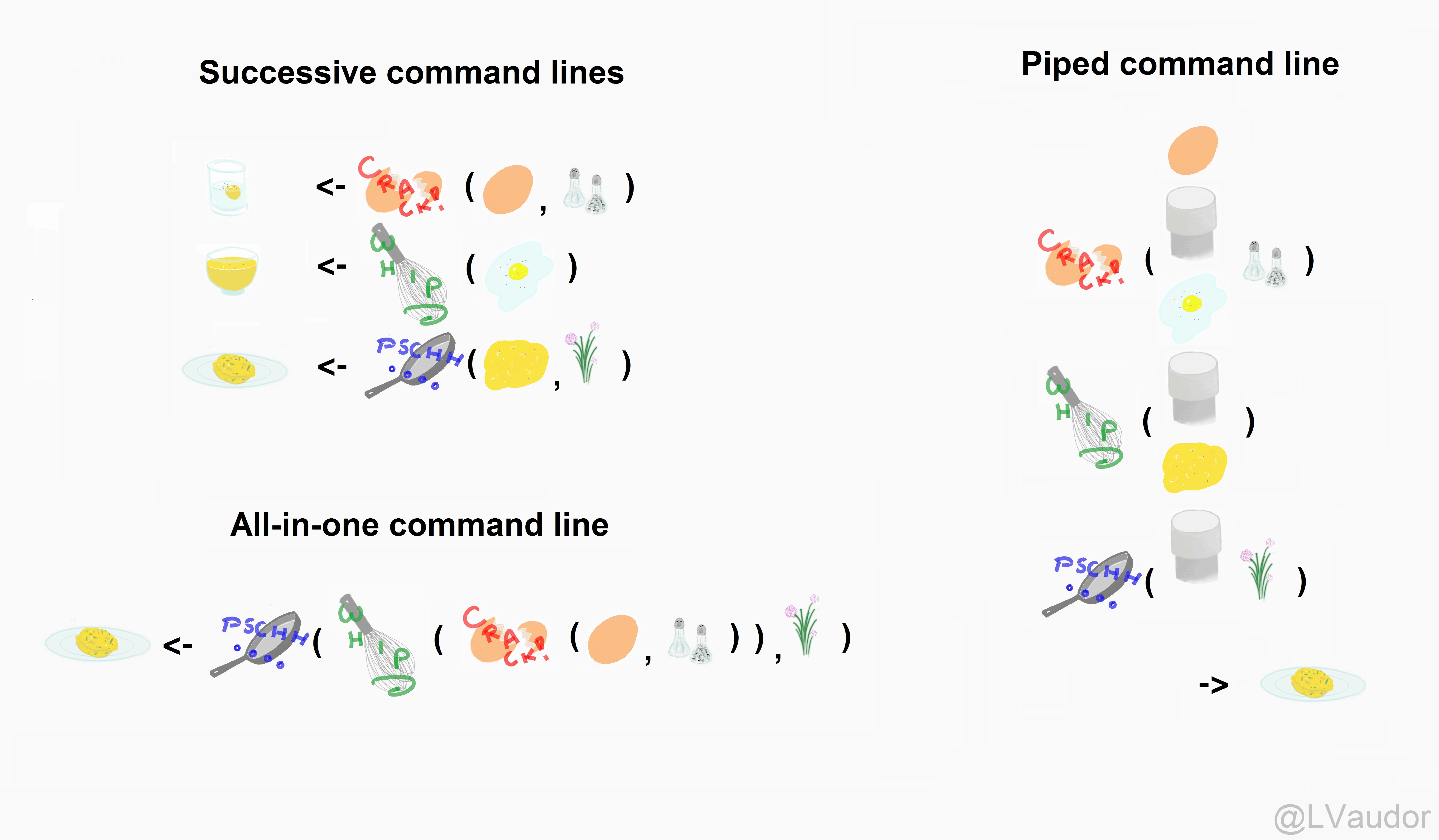
Example from Lise Vaudor
Instead of successive command lines
nlsy2 <- mutate(nlsy, only = case_when(nsibs == 0 ~ "yes", TRUE ~ "no"))nlsy3 <- select(nlsy2, id, contains("sleep"), only)only_kids <- filter(nlsy3, only == "yes")or all-in-one
only_kids <- filter(select(mutate(nlsy, only = case_when(nsibs == 0 ~ "yes", TRUE ~ "no")), id, contains("sleep"), only), only == "yes")It's like reading a story (or nursery rhyme!)
foo_foo <- little_bunny()bop_on( scoop_up( hop_through(foo_foo, forest), field_mouse),head)vs
foo_foo %>% hop_through(forest) %>% scoop_up(field_mouse) %>% bop_on(head)Example from Hadley Wickham

A natural order of operations
leave_house( get_dressed( get_out_of_bed( wake_up(me))))me <- wake_up(me)me <- get_out_of_bed(me)me <- get_dressed(me)me <- leave_house(me)me %>% wake_up() %>% get_out_of_bed() %>% get_dressed() %>% leave_house()
Using pipes with functions we already know
nlsy2 <- mutate(nlsy, only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no"))nlsy3 <- select(nlsy2, id, contains("sleep"), only)only_kids <- filter(nlsy3, only == "yes")only_kids## # A tibble: 30 x 4## id sleep_wkdy sleep_wknd only ## <dbl> <dbl> <dbl> <chr>## 1 458 7 8 yes ## 2 653 6 7 yes ## 3 1101 7 8 yes ## 4 1166 5 6 yes ## 5 2163 7 8 yes ## 6 2442 7 9 yes ## 7 2545 8 8 yes ## 8 3036 5 8 yes ## 9 3194 7 7 yes ## 10 3538 5 5 yes ## # … with 20 more rowsonly_kids <- nlsy %>% mutate(only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no")) %>% select(id, contains("sleep"), only) %>% filter(only == "yes")only_kids## # A tibble: 30 x 4## id sleep_wkdy sleep_wknd only ## <dbl> <dbl> <dbl> <chr>## 1 458 7 8 yes ## 2 653 6 7 yes ## 3 1101 7 8 yes ## 4 1166 5 6 yes ## 5 2163 7 8 yes ## 6 2442 7 9 yes ## 7 2545 8 8 yes ## 8 3036 5 8 yes ## 9 3194 7 7 yes ## 10 3538 5 5 yes ## # … with 20 more rowsPipes replace the first argument of the next function
help(mutate)help(select)help(filter)Usage
mutate(.data, ...)
select(.data, ...)
filter(.data, ...)
Pipes replace the first argument of the next function
nlsy2 <- mutate(nlsy, only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no"))only_kids <- nlsy %>% mutate(only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no"))Pipes replace the first argument of the next function
nlsy2 <- mutate(nlsy, only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no"))nlsy3 <- select(nlsy2, id, contains("sleep"), only)only_kids <- nlsy %>% mutate(only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no")) %>% select(id, contains("sleep"), only)Pipes replace the first argument of the next function
nlsy2 <- mutate(nlsy, only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no"))nlsy3 <- select(nlsy2, id, contains("sleep"), only)only_kids <- filter(nlsy3, only == "yes")only_kids <- nlsy %>% mutate(only = case_when( nsibs == 0 ~ "yes", TRUE ~ "no")) %>% select(id, contains("sleep"), only) %>% filter(only == "yes")1
Your turn...
Exercises 4.1: Try out the pipe!!
Summary statistics
We have seen that we can get certain summary statistics about our data with the summary() function, which we can use either on an entire dataframe/tibble, or on a single variable.
summary(only_kids)## id sleep_wkdy sleep_wknd only ## Min. : 458 Min. :5.000 Min. : 5.000 Length:30 ## 1st Qu.: 3076 1st Qu.:6.000 1st Qu.: 7.000 Class :character ## Median : 4666 Median :7.000 Median : 8.000 Mode :character ## Mean : 5005 Mean :6.833 Mean : 7.633 ## 3rd Qu.: 6823 3rd Qu.:8.000 3rd Qu.: 8.000 ## Max. :12648 Max. :9.000 Max. :12.000summary(nlsy$income)## Min. 1st Qu. Median Mean 3rd Qu. Max. ## 0 6000 11155 15289 20000 75001Summary statistics
We can also apply certain functions to a variable(s) to get a single statistic: mean(), median(), var(), sd(), cov, cor(), min(), max(), quantile(), etc.
median(nlsy$age_bir)## [1] 22cor(nlsy$sleep_wkdy, nlsy$sleep_wknd)## [1] 0.7101579quantile(nlsy$income, probs = c(0.1, 0.9))## 10% 90% ## 3177.2 33024.0Summary statistics
But what if we want a lot of summary statistics -- just not those that come with the summary() function?
- For example, it doesn't give us a standard deviation!
Introducing summarize()
summarize(nlsy, med_age_bir = median(age_bir), cor_sleep = cor(sleep_wkdy, sleep_wknd), ten_pctle_inc = quantile(income, probs = 0.1), ninety_pctle_inc = quantile(income, probs = 0.9))## # A tibble: 1 x 4## med_age_bir cor_sleep ten_pctle_inc ninety_pctle_inc## <dbl> <dbl> <dbl> <dbl>## 1 22 0.710 3177. 33024.summarize() specifics
Usage
summarize(.data, ...)
Arguments
... Name-value pairs of summary functions. The name will be the name of the variable in the result. The value should be an expression that returns a single value like min(x), n(), or sum(is.na(y)).
summarize() specifics
Important to note:
- Takes a dataframe as its first argument. That means we can use pipes!
- Returns a tibble -- helpful if you want to use those values in a figure or table.
- Can give the summary statistics names.
- Can ask for any type of function of the variables (including one you make up yourself).
nlsy %>% summarize(q.1 = quantile(age_bir, probs = 0.1), q.2 = quantile(age_bir, probs = 0.2), q.3 = quantile(age_bir, probs = 0.3), q.4 = quantile(age_bir, probs = 0.4), q.5 = quantile(age_bir, probs = 0.5))## # A tibble: 1 x 5## q.1 q.2 q.3 q.4 q.5## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 17 18 20 21 22Update!
Between my making these slides and now, there was a major update to some functions including summarize()
- You can now provide functions that return multiple values
- But I am keeping the previous example as something we'll work to improve next week!
nlsy %>% summarize(q = quantile(age_bir, seq(from = .01, to = .05, by = .01)), quantile = seq(from = .01, to = .05, by = .01))## # A tibble: 5 x 2## q quantile## <dbl> <dbl>## 1 15 0.01## 2 15 0.02## 3 15 0.03## 4 16 0.04## 5 16 0.05Notice how this output is in rows instead!
Combining summarize with other functions
Because we can pipe, we can also look at statistics of variables that we make using mutate(), in a dataset we've subsetted with filter().
All at once!
nlsy %>% mutate(age_bir_stand = (age_bir - mean(age_bir)) / sd(age_bir)) %>% filter(sex == 1) %>% summarize(mean_men = mean(age_bir_stand))## # A tibble: 1 x 1## mean_men## <dbl>## 1 0.283It's easy to explore your data!
2
Your turn...
Exercises 4.2: Calculate some summary statistics.
What if we want both groups at once?
nlsy %>% filter(sex == 1) %>% summarize(age_bir_men = mean(age_bir))## # A tibble: 1 x 1## age_bir_men## <dbl>## 1 25.1nlsy %>% filter(sex == 2) %>% summarize(age_bir_women = mean(age_bir))## # A tibble: 1 x 1## age_bir_women## <dbl>## 1 22.2We can "group" tibbles using group_by()
We can tell it's "grouped" and how many groups there are by printing out the data.
The data itself won't look different, but we'll be able to perform grouped functions on it.
nlsy_by_region <- group_by(nlsy, region)nlsy_by_region## # A tibble: 1,205 x 14## # Groups: region [4]## glasses eyesight sleep_wkdy sleep_wknd id nsibs samp race_eth sex region income## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0 1 5 7 3 3 5 3 2 1 22390## 2 1 2 6 7 6 1 1 3 1 1 35000## 3 0 2 7 9 8 7 6 3 2 1 7227## 4 1 3 6 7 16 3 5 3 2 1 48000## 5 0 3 10 10 18 2 1 3 1 3 4510## 6 1 2 7 8 20 2 5 3 2 1 50000## 7 0 1 8 8 27 1 5 3 2 1 20000## 8 1 1 8 8 49 6 5 3 2 1 23900## # … with 1,197 more rows, and 3 more variables: res_1980 <dbl>, res_2002 <dbl>,## # age_bir <dbl>group_by()
Like the other functions we've seen, we can use pipes:
nlsy %>% mutate(income_stand = (income - mean(income))/sd(income)) %>% select(id, region, income_stand, race_eth, sex) %>% group_by(race_eth)## # A tibble: 1,205 x 5## # Groups: race_eth [3]## id region income_stand race_eth sex## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 3 1 0.533 3 2## 2 6 1 1.48 3 1## 3 8 1 -0.605 3 2## 4 16 1 2.45 3 2## 5 18 3 -0.809 3 1## 6 20 1 2.60 3 2## 7 27 1 0.353 3 2## 8 49 1 0.646 3 2## # … with 1,197 more rowsStratify with group_by() %>% summarize()
This function is especially important when calculating summary statistics, which we often want to be stratified.
nlsy %>% mutate(income_stand = (income - mean(income))/sd(income)) %>% group_by(region) %>% summarize(mean_inc = mean(income_stand), sd_inc = sd(income_stand))## # A tibble: 4 x 3## region mean_inc sd_inc## <dbl> <dbl> <dbl>## 1 1 0.186 1.17 ## 2 2 0.106 0.958## 3 3 -0.0891 1.03 ## 4 4 -0.145 0.810Multiple layers of groups
We can group by multiple variables:
nlsy %>% group_by(region, sex) %>% summarize(mean_inc = mean(income), sd_inc = sd(income))## # A tibble: 8 x 4## # Groups: region [4]## region sex mean_inc sd_inc## <dbl> <dbl> <dbl> <dbl>## 1 1 1 20393. 16929.## 2 1 2 15928. 14442.## 3 2 1 19488. 14885.## 4 2 2 14699. 10613.## 5 3 1 15137. 14592.## 6 3 2 13293. 12879.## 7 4 1 12315. 9244.## 8 4 2 14001. 11621.This would be much easier to read if we had made region and sex into factor variables with labels!
Counting groups
Sometimes we just want to know how many observations are in a group. We can do that (at least) three ways!
nlsy %>% group_by(sex) %>% summarize(n = n())## # A tibble: 2 x 2## sex n## <dbl> <int>## 1 1 501## 2 2 704nlsy %>% group_by(sex) %>% tally()## # A tibble: 2 x 2## sex n## <dbl> <int>## 1 1 501## 2 2 704nlsy %>% count(sex)## # A tibble: 2 x 2## sex n## <dbl> <int>## 1 1 501## 2 2 704Calculating proportions
We can add an extra step to calculate proportions.
- Group by the variable for which we want proportions
- Count the number of observations in each group
- Divide that number by the sum of the counts across all groups
nlsy %>% group_by(sex) %>% summarize(n = n()) %>% mutate(prop = n / sum(n))## # A tibble: 2 x 3## sex n prop## <dbl> <int> <dbl>## 1 1 501 0.416## 2 2 704 0.5843
Your turn...
Exercises 4.3: Calculate grouped statistics.
Making a table 1
Almost every study will involve a Table 1, where you present summary statistics about your sample, often stratified.
There are a number of R packages that can help make your life easier and prevent errors.
tableone package
install.packages("tableone")library(tableone)tab1 <- CreateTableOne( data = nlsy, vars = c("eyesight", "nsibs", "race_eth", "sex", "region", "income", "age_bir"), strata = "glasses", factorVars = c("eyesight", "race_eth", "sex", "region"))tab1## Stratified by glasses## 0 1 p test## n 581 624 ## eyesight (%) 0.333 ## 1 220 (37.9) 254 (40.7) ## 2 185 (31.8) 200 (32.1) ## 3 122 (21.0) 127 (20.4) ## 4 46 ( 7.9) 32 ( 5.1) ## 5 8 ( 1.4) 11 ( 1.8) ## nsibs (mean (SD)) 4.03 (2.65) 3.85 (2.47) 0.210 ## race_eth (%) <0.001 ## 1 103 (17.7) 108 (17.3) ## 2 177 (30.5) 130 (20.8) ## 3 301 (51.8) 386 (61.9) ## sex = 2 (%) 301 (51.8) 403 (64.6) <0.001 ## region (%) 0.230 ## 1 102 (17.6) 104 (16.7) ## 2 146 (25.1) 187 (30.0) ## 3 211 (36.3) 200 (32.1) ## 4 122 (21.0) 133 (21.3) ## income (mean (SD)) 13852.78 (12011.51) 16626.22 (14329.16) <0.001 ## age_bir (mean (SD)) 23.07 (5.91) 23.80 (6.05) 0.036tableone options
print(tab1, catDigits = 2, contDigits = 2, test = FALSE, smd = TRUE)## Stratified by glasses## 0 1 SMD ## n 581 624 ## eyesight (%) 0.123## 1 220 (37.87) 254 (40.71) ## 2 185 (31.84) 200 (32.05) ## 3 122 (21.00) 127 (20.35) ## 4 46 ( 7.92) 32 ( 5.13) ## 5 8 ( 1.38) 11 ( 1.76) ## nsibs (mean (SD)) 4.03 (2.65) 3.85 (2.47) 0.072## race_eth (%) 0.234## 1 103 (17.73) 108 (17.31) ## 2 177 (30.46) 130 (20.83) ## 3 301 (51.81) 386 (61.86) ## sex = 2 (%) 301 (51.81) 403 (64.58) 0.261## region (%) 0.120## 1 102 (17.56) 104 (16.67) ## 2 146 (25.13) 187 (29.97) ## 3 211 (36.32) 200 (32.05) ## 4 122 (21.00) 133 (21.31) ## income (mean (SD)) 13852.78 (12011.51) 16626.22 (14329.16) 0.210## age_bir (mean (SD)) 23.07 (5.91) 23.80 (6.05) 0.121tableone resources
See all the options:
help("CreateTableOne")help("print.TableOne")You can read a walk-through of package options here: https://cran.r-project.org/web/packages/tableone/vignettes/introduction.html
You can see a list of similar packages here: https://github.com/kaz-yos/tableone#similar-or-complementary-projects
4
Your turn...
Exercises 4.4: Try out the Table 1 package!