Introduction to R
Week 5: Functions
Louisa Smith
August 10 - August 14
Let's be
efficient
Previously...
analysis_dat <- nlsy %>% mutate(ineligible = case_when( income > 50000 ~ 1, age_bir > 35 ~ 1, TRUE ~ 0 )) %>% filter(ineligible == 0) %>% select(id, sex, race_eth, glasses, eyesight)analysis_dat## # A tibble: 1,128 x 5## id sex race_eth glasses eyesight## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 3 2 3 0 1## 2 6 1 3 1 2## 3 8 2 3 0 2## 4 16 2 3 1 3## 5 18 1 3 0 3## 6 20 2 3 1 2## 7 27 2 3 0 1## # … with 1,121 more rowsPreviously...
stats <- analysis_dat %>% mutate(sex = factor(sex, labels = c("male", "female")), race_eth = factor(race_eth, labels = c("Hispanic", "Black", "Other"))) %>% group_by(race_eth, sex) %>% summarize(prop_glass = mean(glasses), sd_eyesight = sd(eyesight))stats## # A tibble: 6 x 4## # Groups: race_eth [3]## race_eth sex prop_glass sd_eyesight## <fct> <fct> <dbl> <dbl>## 1 Hispanic male 0.403 0.894## 2 Hispanic female 0.566 1.10 ## 3 Black male 0.318 0.971## 4 Black female 0.488 1.11 ## 5 Other male 0.490 0.941## 6 Other female 0.602 0.972Previously...
ggplot(stats) + geom_col(aes(x = sex, y = prop_glass, fill = sex)) + facet_grid(cols = vars(race_eth)) + scale_fill_brewer(palette = "Set1", guide = "none") + theme_minimal() + labs(x = NULL, y = "proportion wearing glasses")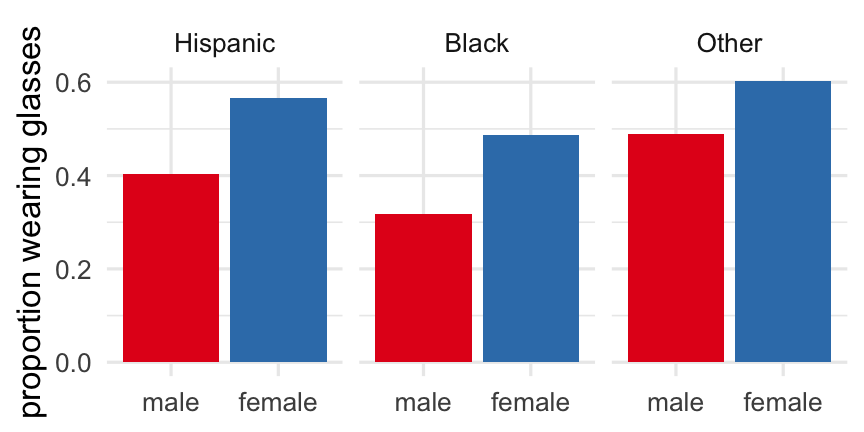
Previously...
tab1 <- CreateTableOne( data = analysis_dat, strata = "sex", vars = c("race_eth", "glasses", "eyesight"), factorVars = c("race_eth", "glasses"))print(tab1, test = FALSE, catDigits = 0, contDigits = 0)## Stratified by sex## 1 2 ## n 453 675 ## race_eth (%) ## 1 77 (17) 129 (19) ## 2 129 (28) 164 (24) ## 3 247 (55) 382 (57) ## glasses = 1 (%) 193 (43) 383 (57) ## eyesight (mean (SD)) 2 (1) 2 (1)Functions in R
I've been denoting functions with parentheses: func()
We've seen functions such as:
mean()theme_minimal()mutate()case_when()group_by()CreateTableOne()
Functions take arguments and return values
Looking inside a function
If you want to see the code within a function, you can just type its name without the parentheses:
CreateTableOne## function (vars, strata, data, factorVars, includeNA = FALSE, ## test = TRUE, testApprox = chisq.test, argsApprox = list(correct = TRUE), ## testExact = fisher.test, argsExact = list(workspace = 2 * ## 10^5), testNormal = oneway.test, argsNormal = list(var.equal = TRUE), ## testNonNormal = kruskal.test, argsNonNormal = list(NULL), ## smd = TRUE, addOverall = FALSE) ## {## ModuleStopIfNotDataFrame(data)## if (missing(vars)) {## vars <- names(data)## }## vars <- ModuleReturnVarsExist(vars, data)## ModuleStopIfNoVarsLeft(vars)## varLabels <- labelled::var_label(data[vars])## if (!missing(factorVars)) {## factorVars <- ModuleReturnVarsExist(factorVars, data)## data[factorVars] <- lapply(data[factorVars], factor)## }## test <- ModuleReturnFalseIfNoStrata(strata, test)## smd <- ModuleReturnFalseIfNoStrata(strata, smd)## percentMissing <- ModulePercentMissing(data[vars])## varClasses <- lapply(data[vars], class)## varFactors <- sapply(varClasses, function(VEC) {## any(VEC %in% c("factor", "ordered", "logical", "character"))## })## varFactors <- names(varFactors)[varFactors]## varNumerics <- sapply(varClasses, function(VEC) {## any(VEC %in% c("numeric", "integer", "double"))## })## varNumerics <- names(varNumerics)[varNumerics]## varDrop <- setdiff(vars, c(varFactors, varNumerics))## if (length(varDrop) > 0) {## warning("Dropping variable(s) ", paste0(varDrop, sep = " "), ## " due to unsupported class.\n")## vars <- setdiff(vars, varDrop)## }## logiFactors <- vars %in% varFactors## argsCreateContTable <- list(data = data, test = test, testNormal = testNormal, ## argsNormal = argsNormal, testNonNormal = testNonNormal, ## argsNonNormal = argsNonNormal, smd = smd, addOverall = addOverall)## argsCreateCatTable <- list(data = data, includeNA = includeNA, ## test = test, testApprox = testApprox, argsApprox = argsApprox, ## testExact = testExact, argsExact = argsExact, smd = smd, ## addOverall = addOverall)## if (!missing(strata)) {## dfStrata <- ModuleReturnStrata(strata, data)## strata <- names(dfStrata)## argsCreateContTable <- c(list(strata = strata), argsCreateContTable)## argsCreateCatTable <- c(list(strata = strata), argsCreateCatTable)## }## if (length(varNumerics) == 0) {## ContTable <- NULL## CatTable <- do.call(CreateCatTable, args = c(list(vars = varFactors), ## argsCreateCatTable))## }## else if (length(varFactors) == 0) {## ContTable <- do.call(CreateContTable, args = c(list(vars = varNumerics), ## argsCreateContTable))## CatTable <- NULL## }## else if ((length(varFactors) > 0) & (length(varNumerics) > ## 0)) {## ContTable <- do.call(CreateContTable, args = c(list(vars = varNumerics), ## argsCreateContTable))## CatTable <- do.call(CreateCatTable, args = c(list(vars = varFactors), ## argsCreateCatTable))## }## else {## warning("No variables left to analyzed in vars.")## }## TableOneObject <- list(ContTable = ContTable, CatTable = CatTable, ## MetaData = list(vars = vars, logiFactors = logiFactors, ## varFactors = varFactors, varNumerics = varNumerics, ## percentMissing = percentMissing, varLabels = varLabels))## class(TableOneObject) <- "TableOne"## return(TableOneObject)## }## <bytecode: 0x7f7f75edfdc8>## <environment: namespace:tableone>Structure of a function
func <- function()You can name your function like you do any other object
- Just avoid names of existing functions
Structure of a function
func <- function(arg1, arg2 = default_val)}What objects/values do you need to make your function work?
- You can give them default values to use if the user doesn't specify others
Structure of a function
func <- function(arg1, arg2 = default_val) {}Everything else goes within curly braces
- Code in here will essentially look like any other R code, using any inputs to your functions
Structure of a function
func <- function(arg1, arg2 = default_val) { new_val <- # do something with args}Make new objects
One thing you'll likely want to do is make new objects along the way
These aren't saved to your environment (i.e., you won't see them in the upper-right window) when you run the function
You can think of them as being stored in a temporary environment within the function
Structure of a function
func <- function(arg1, arg2 = default_val) { new_val <- # do something with args return(new_val)}Return something new that the code has produced
- The
return()statement is actually optional. If you don't put it, it will return the last object in the code. When you're starting out, it's safer to always explicitly write out what you want to return.
Example: a new function for the mean
Let's say we are not satisfied with the mean() function and want to write our own.
Here's the general structure we'll start with.
new_mean <- function() {}New mean: arguments
We'll want to take the mean of a vector of numbers.
It will help to make an example of such a vector to think about what the input might look like, and to test the function. We'll call it x:
x <- c(1, 3, 5, 7, 9)We can add x as an argument to our function:
new_mean <- function(x) {}New mean: function body
Let's think about how we calculate a mean in math, and then translate it into code:
¯x=1nn∑i=1xi
So we need to sum the elements of x together, and then divide by the number of elements.
New mean: function body
Let's think about how we calculate a mean in math, and then translate it into code:
¯x=1nn∑i=1xi
So we need to sum the elements of x together, and then divide by the number of elements.
We can use the functions sum() and length() to help us.
We'll write the code with our test vector first, before inserting it into the function:
n <- length(x)sum(x) / n## [1] 5New mean: function body
Our code seems to be doing what we want, so let's insert it. To be explicit, I've stored the answer (within the function) as mean_val, then returned that value.
new_mean <- function(x) { n <- length(x) mean_val <- sum(x) / n return(mean_val)}Testing a function
Let's plug in the vector that we created to test it:
new_mean(x = x)## [1] 5And then try another one we create on the spot:
new_mean(x = c(100, 200, 300))## [1] 200Great!
Adding another argument
Let's say we plan to be using our new_mean() function to calculate proportions (i.e., the mean of a binary variable). Sometimes we'll want to report them as multiplier by multiplying the proportion by 100.
Let's name our new function prop(). We'll use the same structure as we did with new_mean().
prop <- function(x) { n <- length(x) mean_val <- sum(x) / n return(mean_val)}Testing the code
Now we'll want to test on a vector of 1's and 0's.
x <- c(0, 1, 1)To calculate the proportion and turn it into a percentage, we'll just multiply the mean by 100.
multiplier <- 100multiplier * sum(x) / length(x)## [1] 66.66667Testing the code
We want to give users the option to choose between a proportion and a percentage. So we'll add an argument multiplier. When we want to just return the proportion, we can just set multiplier to be 1.
multiplier <- 1multiplier * sum(x) / length(x)## [1] 0.6666667multiplier <- 100multiplier * sum(x) / length(x)## [1] 66.66667Adding another argument
If we add multiplier as an argument, we can refer to it in the function body.
prop <- function(x, multiplier) { n <- length(x) mean_val <- multiplier * sum(x) / n return(mean_val)}Adding another argument
Now we can test:
prop(x = c(1, 0, 1, 0), multiplier = 1)## [1] 0.5prop(x = c(1, 0, 1, 0), multiplier = 100)## [1] 50Making a default argument
Since we don't want users to have to specify multiplier = 1 every time they just want a proportion, we can set it as a default.
prop <- function(x, multiplier = 1) { n <- length(x) mean_val <- multiplier * sum(x) / n return(mean_val)}Now we only need to specify that argument if we want a percentage.
prop(x = c(0, 1, 1, 1))## [1] 0.75prop(x = c(0, 1, 1, 1), multiplier = 100)## [1] 75Caveats
This is obviously not the best way to write this function!
For example, it will still work if
x = c(123, 593, -192).... but it certainly won't give you a proportion or a percentage!We could also put
multiplier =any number, and we'll just be multiplying the answer by that number -- this is essentially meaningless.We also haven't done any checking to see whether the user is even entering numbers! We could put in better error messages so users don't just get an R default error message if they do something wrong.
prop(x = c("blah", "blah", "blah"))## Error in sum(x): invalid 'type' (character) of argument1
Your turn...
Exercises 5.1: Write a function to calculate exponents.
When to make a function
There's a rule somewhere that says that if you are copying and pasting something 3 times in your code, you should just make a function to do it instead.
For example, when we were calculating quantiles:
nlsy %>% summarize(q.1 = quantile(age_bir, probs = 0.1), q.2 = quantile(age_bir, probs = 0.2), q.3 = quantile(age_bir, probs = 0.3), q.4 = quantile(age_bir, probs = 0.4), q.5 = quantile(age_bir, probs = 0.5))## # A tibble: 1 x 5## q.1 q.2 q.3 q.4 q.5## <dbl> <dbl> <dbl> <dbl> <dbl>## 1 17 18 20 21 22We could make a function to do this instead!
Age at first birth quantile function
What will our argument(s) be? How about just the quantile of interest, to start out, which we can refer to as q.
What will the name of our function be? Since we're looking at quantiles of age at first birth, let's call it age_bir_q():
age_bir_q <- function(q) {}Prepare the code
First let's choose a value to help us write the code for the body of our function:
q <- .5Then we can write the code with reference to the variable q.
nlsy %>% summarize( q_var = quantile(age_bir, probs = q) )## # A tibble: 1 x 1## q_var## <dbl>## 1 22Copy and paste just once
age_bir_q <- function(q) { quant <- nlsy %>% summarize(q_var = quantile(age_bir, probs = q)) return(quant)}It's always good to check your function, if possible, with some other way to get the same result. Here we can double check using the median:
age_bir_q(q = 0.5)## # A tibble: 1 x 1## q_var## <dbl>## 1 22median(nlsy$age_bir)## [1] 22What if we want to change the variable
This is where things get a little tricky. It's hard to use an unquoted variable name as an argument to a function. Since it's not an object in the environment, R will complain if we try to do something like this:
var_q <- function(q, var) { quant <- nlsy %>% summarize(q_var = quantile(var, probs = q)) return(quant)}var_q(q = 0.5, var = income)## Error: Problem with summarise() input q_var.## x object 'income' not found## ℹ Input q_var is quantile(var, probs = q).We might think it would help if we put income in quotes, but alas!
var_q(q = 0.5, var = "income")## Error: Problem with summarise() input q_var.## x non-numeric argument to binary operator## ℹ Input q_var is quantile(var, probs = q).What if we want to change the variable
There are more "official" ways to deal with this that are beyond the scope of this class, but there's usually a workaround to be able to write your variable name as a character string instead.
Consider that we can rename a variable using the rename() function, which is a function that can take variable names in quotes:
nlsy %>% rename(eyeglasses = "glasses")## # A tibble: 1,205 x 10## eyeglasses eyesight sleep_wkdy sleep_wknd id nsibs samp race_eth sex region## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0 1 5 7 3 3 5 3 2 1## 2 1 2 6 7 6 1 1 3 1 1## 3 0 2 7 9 8 7 6 3 2 1## 4 1 3 6 7 16 3 5 3 2 1## 5 0 3 10 10 18 2 1 3 1 3## 6 1 2 7 8 20 2 5 3 2 1## 7 0 1 8 8 27 1 5 3 2 1## # … with 1,198 more rowsWhat if we want to change the variable
Let's just rename the variable we want to new_var, then we can pass the variable new_var to any function we want:
var_q <- function(q, var) { quant <- nlsy %>% rename(new_var = var) %>% summarize(q_var = quantile(new_var, probs = q)) return(quant)}var_q(q = 0.5, var = "income")## # A tibble: 1 x 1## q_var## <dbl>## 1 11155Use our function on any combination of var and q
var_q(q = 0.25, var = "sleep_wkdy")## # A tibble: 1 x 1## q_var## <dbl>## 1 6Use our function on any combination of var and q
var_q(q = 0.95, var = "nsibs")## # A tibble: 1 x 1## q_var## <dbl>## 1 9Changing a grouping variable
We might run into the same problem with wanting to change a variable, if, say, we want to calculate the mean for a number of different variables:
nlsy %>% group_by(sex) %>% summarize(mean_inc = mean(income))## # A tibble: 2 x 2## sex mean_inc## <dbl> <dbl>## 1 1 16690.## 2 2 14292.nlsy %>% group_by(glasses) %>% summarize(mean_inc = mean(income))## # A tibble: 2 x 2## glasses mean_inc## <dbl> <dbl>## 1 0 13853.## 2 1 16626.It will be your job in the exercises to write a function to do this!
2
Your turn...
Exercises 5.2: Write a function to calculate grouped means.
Repeating functions
Often we want to repeat functions, or some procedure, over and over again.
One option which you may be familiar with from other programming languages is a for loop:
for (i in 1:3) { print(i)}## [1] 1## [1] 2## [1] 3Structure of a for loop
for (i in vals) { something(i) # do things here!}If we want to print our results to the console, we have to use the print() function
qs <- c(0.1, 0.5, 0.9)for (i in qs) { print(var_q(q = i, var = "income"))}## # A tibble: 1 x 1## q_var## <dbl>## 1 3177.## # A tibble: 1 x 1## q_var## <dbl>## 1 11155## # A tibble: 1 x 1## q_var## <dbl>## 1 33024.If we want to save our results, we should set up an empty object to do so
results <- rep(NA, 3)for (i in 1:3) { results[[i]] <- i * 1.5}results## [1] 1.5 3.0 4.5What just happened?
results <- rep(NA, 3)results # empty vector of NAs## [1] NA NA NAfor (i in 1:3) { # fill the i'th entry with # the value i times 1.5 results[[i]] <- i * 1.5 }results## [1] 1.5 3.0 4.5Quick detour back to our function
Let's return just the q_var column, not the whole tibble that was created (since this function is really just calculating one number)
- The
pull()function pulls off a vector; in this case, one number
var_q_new <- function(q, var) { quant <- nlsy %>% rename(new_var = var) %>% summarize(q_var = quantile(new_var, probs = q)) %>% pull(q_var) return(quant)}var_q_new(q = 0.5, var = "income")## 50% ## 11155If we want to calculate all the deciles of income
We're going to want to calculate 10 values for 10 different arguments to quantile.
- Here are the arguments we're going to cycle through:
# use seq to generate values from# 0.1 to 0.9, skipping along by 0.1qs <- seq(0.1, 0.9, by = 0.1)qs## [1] 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9How are we going to cycle through qs?
The seq_along function is the best way to go from 1 to the length of your vector:
seq_along(qs)## [1] 1 2 3 4 5 6 7 8 9We can extract the value from qs that we want with whatever value i is at:
i <- 4 # (for example)qs[[i]]## [1] 0.4Where are we going to store these 10 numbers?
# use length() to get the right number of# empty values without even thinking!deciles <- rep(NA, length(qs))deciles## [1] NA NA NA NA NA NA NA NA NAWe can access these with i as well:
deciles[[i]] # 4th NA, since i is currently 4## [1] NAPutting it all together
for (i in seq_along(qs)) { deciles[[i]] <- var_q_new(q = qs[[i]], var = "income")}deciles## [1] 3177.2 5025.6 6907.2 9000.0 11155.0 14000.0 18053.6 23800.0 33024.0Notes on for loops
The
iis arbitrary... you can cycle through whatever variable you want, you don't have to call iti!People may try to tell you that for loops in R are slow. This is generally only true if you don't make an empty vector or matrix to hold your results ahead of time.
That said, there's often a more concise and readable equivalent to a for loop in R. The
apply()family of functions is one option (brief guide here), but I have started exclusively using thepurrrpackage and itsmap()family. The "iteration" chapter in the R for Data Science book is highly recommended.
3
Your turn...
Exercises 5.3: Write a for loop.
Other options
This class introduced you to the basics... but there are usually easier/more efficient ways to do everything. I'll show you some examples of some more advanced topics that you can look more into on your own.
- You'll know they exist, so come back when you're ready for them!
Summarize multiple variables with multiple functions
nlsy %>% summarize(across(contains("sleep"), list(med = median, sd = sd)))## # A tibble: 1 x 4## sleep_wkdy_med sleep_wkdy_sd sleep_wknd_med sleep_wknd_sd## <dbl> <dbl> <dbl> <dbl>## 1 7 1.34 7 1.50Summarize all numeric variables at once
nlsy %>% summarize(across(where(is.numeric), mean))## # A tibble: 1 x 14## glasses eyesight sleep_wkdy sleep_wknd id nsibs samp race_eth sex region income## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0.518 1.99 6.64 7.27 5229. 3.94 7.00 2.40 1.58 2.59 15289.## # … with 3 more variables: res_1980 <dbl>, res_2002 <dbl>, age_bir <dbl>Make a series of consecutive variables factors
nlsy %>% mutate(across(c(race_eth:region), factor))## # A tibble: 1,205 x 14## glasses eyesight sleep_wkdy sleep_wknd id nsibs samp race_eth sex region income## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <fct> <fct> <dbl>## 1 0 1 5 7 3 3 5 3 2 1 22390## 2 1 2 6 7 6 1 1 3 1 1 35000## 3 0 2 7 9 8 7 6 3 2 1 7227## 4 1 3 6 7 16 3 5 3 2 1 48000## 5 0 3 10 10 18 2 1 3 1 3 4510## 6 1 2 7 8 20 2 5 3 2 1 50000## 7 0 1 8 8 27 1 5 3 2 1 20000## 8 1 1 8 8 49 6 5 3 2 1 23900## 9 1 2 7 8 57 1 5 3 2 1 23289## 10 0 1 8 8 67 1 1 3 1 1 35000## # … with 1,195 more rows, and 3 more variables: res_1980 <dbl>, res_2002 <dbl>,## # age_bir <dbl>Rename all your variables
nlsy %>% rename_with(toupper)## # A tibble: 1,205 x 14## GLASSES EYESIGHT SLEEP_WKDY SLEEP_WKND ID NSIBS SAMP RACE_ETH SEX REGION INCOME## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>## 1 0 1 5 7 3 3 5 3 2 1 22390## 2 1 2 6 7 6 1 1 3 1 1 35000## 3 0 2 7 9 8 7 6 3 2 1 7227## 4 1 3 6 7 16 3 5 3 2 1 48000## 5 0 3 10 10 18 2 1 3 1 3 4510## 6 1 2 7 8 20 2 5 3 2 1 50000## 7 0 1 8 8 27 1 5 3 2 1 20000## 8 1 1 8 8 49 6 5 3 2 1 23900## 9 1 2 7 8 57 1 5 3 2 1 23289## 10 0 1 8 8 67 1 1 3 1 1 35000## # … with 1,195 more rows, and 3 more variables: RES_1980 <dbl>, RES_2002 <dbl>,## # AGE_BIR <dbl>Resources
- Vignette about applying function to multiple columns
- Vignette about summarizing across rows
- Two videos about some more advanced topics that allow us to pass variable names to functions: https://www.youtube.com/watch?v=nERXS3ssntw and https://www.youtube.com/watch?v=2-gknoyjL3A
- Blog post on the
apply()family of functions: https://petewerner.blogspot.com/2012/12/using-apply-sapply-lapply-in-r.html - Video tutorial on the
map()family of functions: https://resources.rstudio.com/wistia-rstudio-conf-2017/happy-r-users-purrr-tutorial-charlotte-wickham
4
Your turn...
Exercises 5.4: Choose one of the resources -- from any of the lectures -- that you haven't looked through yet and read through it.