Lab
6 Functions
You can either download the lab as an RMarkdown file here, or copy and paste the code as we go into a .R script. Either way, save it into the 06-week folder where you completed the exercises! Since we worked on file structure this week, put it in a new labs folder nested within that folder.
6.1 File paths
We talked this week about making file paths work in R. This is especially important if you’re sharing your code with everyone. If you set your working directory via setwd() or hard-code another path into your code, it’s definitely not going to work on my computer.
A famous blog post about this (well, as famous as blog posts about R can be) can be read here.
How did you make these file paths work?
library(tidyverse)
library(knitr)
nlsy <- read_csv("nlsy.csv")# this function adds a static image
include_graphics("figure1.jpg")read_rds("table1.rds") %>%
kable() # this function prints nicer tables in R MarkdownBy the way, you can change the directory in which an R Markdown document is knitted using the arrow by the Knit button. You can also use the settings to change the default. But if you use the here package it won’t matter what you do!
One thing that is helpful to know when you’re working interactively, is how to navigate around file paths. If you need to go up a level, you use two dots to do so. So in the code below, we go down two levels from the top-level directory, then have to go up two levels and down two different levels to access the data. See why project directories and the here package make everything easier?!
proj_wd <- getwd()
setwd("figures/super important")
nlsy <- read_csv("../../data/raw/nlsy.csv")
# ahhh take us back to where we started from!
setwd(proj_wd)6.2 Missing data
The nlsy.csv file includes all 12,687 participants of the NLSY-79. Read in the data once without specifying the values that indicate missingness. Explore the data and find them all. Then read in the data again, using the na = argument in read_csv() to read them in as NA’s.
# skip = 1 means to skip the first row, which were the original col names
nlsy <- read_csv(here::here("data", "raw", "nlsy.csv"), skip = 1, col_names = c(
"glasses", "eyesight", "sleep_wkdy", "sleep_wknd",
"id", "nsibs", "samp", "race_eth", "sex", "region",
"income", "res_1980", "res_2002", "age_bir"
))How can we explore the data to get an idea of what values mean that variable is missing?
summary(nlsy)#> glasses eyesight sleep_wkdy sleep_wknd
#> Min. :-4.000 Min. :-4.00000 Min. :-4.000 Min. :-4.000
#> 1st Qu.:-4.000 1st Qu.:-4.00000 1st Qu.:-4.000 1st Qu.:-4.000
#> Median : 0.000 Median : 1.00000 Median : 6.000 Median : 6.000
#> Mean :-1.026 Mean : 0.04004 Mean : 2.523 Mean : 2.886
#> 3rd Qu.: 1.000 3rd Qu.: 2.00000 3rd Qu.: 7.000 3rd Qu.: 8.000
#> Max. : 1.000 Max. : 5.00000 Max. :19.000 Max. :24.000
#> id nsibs samp race_eth
#> Min. : 1 Min. :-3.000 Min. : 1.000 Min. :1.000
#> 1st Qu.: 3172 1st Qu.: 2.000 1st Qu.: 5.000 1st Qu.:2.000
#> Median : 6344 Median : 3.000 Median : 9.000 Median :3.000
#> Mean : 6344 Mean : 3.844 Mean : 8.174 Mean :2.434
#> 3rd Qu.: 9515 3rd Qu.: 5.000 3rd Qu.:12.000 3rd Qu.:3.000
#> Max. :12686 Max. :29.000 Max. :20.000 Max. :3.000
#> sex region income res_1980
#> Min. :1.000 Min. :-4.000 Min. : -3 Min. :-5.0000
#> 1st Qu.:1.000 1st Qu.: 2.000 1st Qu.: 2853 1st Qu.:-4.0000
#> Median :1.000 Median : 3.000 Median : 7955 Median :-4.0000
#> Mean :1.495 Mean : 2.424 Mean :11819 Mean : 0.5317
#> 3rd Qu.:2.000 3rd Qu.: 3.000 3rd Qu.:18000 3rd Qu.:11.0000
#> Max. :2.000 Max. : 4.000 Max. :75001 Max. :16.0000
#> res_2002 age_bir
#> Min. :-5.000 Min. :-998.00
#> 1st Qu.:-5.000 1st Qu.: -5.00
#> Median :11.000 Median : -5.00
#> Mean : 4.914 Mean : -96.36
#> 3rd Qu.:11.000 3rd Qu.: 23.00
#> Max. :19.000 Max. : 52.00ggplot(nlsy) +
geom_histogram(aes(age_bir))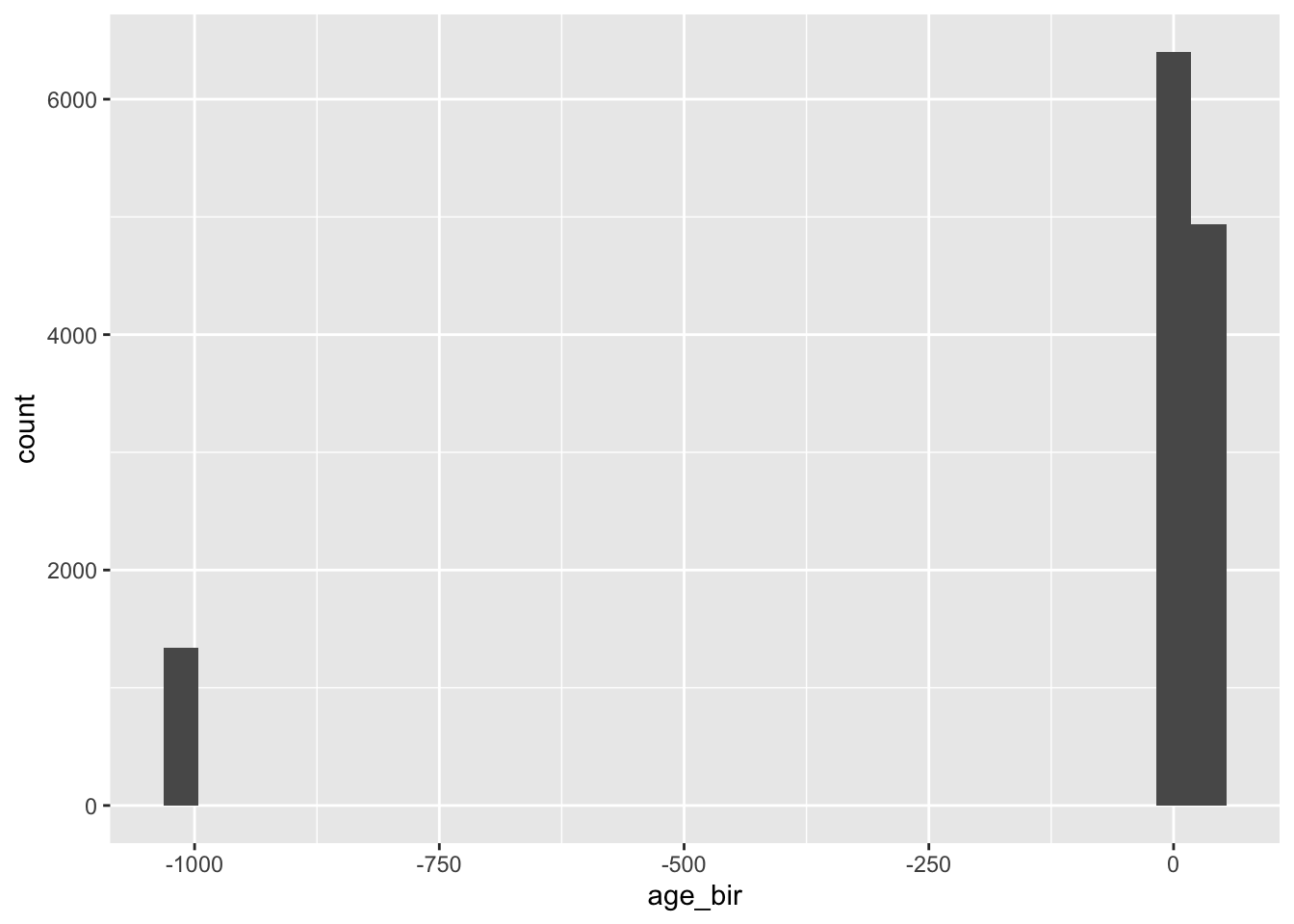
count(nlsy, nsibs)#> # A tibble: 25 x 2
#> nsibs n
#> <dbl> <int>
#> 1 -3 14
#> 2 -2 3
#> 3 -1 1
#> 4 0 362
#> 5 1 1611
#> 6 2 2530
#> 7 3 2412
#> 8 4 1783
#> 9 5 1198
#> 10 6 933
#> # … with 15 more rowsReread the data with the missing values specified:
nlsy <- read_csv(here::here("data", "raw", "nlsy.csv"), skip = 1,
na = c("-998", "-5", "-4", "-3", "-2", "-1"),
col_names = c(
"glasses", "eyesight", "sleep_wkdy", "sleep_wknd",
"id", "nsibs", "samp", "race_eth", "sex", "region",
"income", "res_1980", "res_2002", "age_bir"
))Check again to see if anything’s off:
summary(nlsy)#> glasses eyesight sleep_wkdy sleep_wknd
#> Min. :0.000 Min. :1.000 Min. : 0.000 Min. : 0.000
#> 1st Qu.:0.000 1st Qu.:1.000 1st Qu.: 6.000 1st Qu.: 6.000
#> Median :0.000 Median :2.000 Median : 7.000 Median : 7.000
#> Mean :0.461 Mean :2.064 Mean : 6.567 Mean : 7.163
#> 3rd Qu.:1.000 3rd Qu.:3.000 3rd Qu.: 8.000 3rd Qu.: 8.000
#> Max. :1.000 Max. :5.000 Max. :19.000 Max. :24.000
#> NA's :4236 NA's :4242 NA's :4860 NA's :4867
#> id nsibs samp race_eth
#> Min. : 1 Min. : 0.000 Min. : 1.000 Min. :1.000
#> 1st Qu.: 3172 1st Qu.: 2.000 1st Qu.: 5.000 1st Qu.:2.000
#> Median : 6344 Median : 3.000 Median : 9.000 Median :3.000
#> Mean : 6344 Mean : 3.854 Mean : 8.174 Mean :2.434
#> 3rd Qu.: 9515 3rd Qu.: 5.000 3rd Qu.:12.000 3rd Qu.:3.000
#> Max. :12686 Max. :29.000 Max. :20.000 Max. :3.000
#> NA's :18
#> sex region income res_1980
#> Min. :1.000 Min. :1.000 Min. : 0 Min. : 1.000
#> 1st Qu.:1.000 1st Qu.:2.000 1st Qu.: 5900 1st Qu.: 6.000
#> Median :1.000 Median :3.000 Median :10700 Median :11.000
#> Mean :1.495 Mean :2.547 Mean :14707 Mean : 8.896
#> 3rd Qu.:2.000 3rd Qu.:3.000 3rd Qu.:20000 3rd Qu.:11.000
#> Max. :2.000 Max. :4.000 Max. :75001 Max. :16.000
#> NA's :239 NA's :2491 NA's :8186
#> res_2002 age_bir
#> Min. : 1.00 Min. :10.00
#> 1st Qu.:11.00 1st Qu.:20.00
#> Median :11.00 Median :23.00
#> Mean :11.28 Mean :24.38
#> 3rd Qu.:11.00 3rd Qu.:28.00
#> Max. :19.00 Max. :52.00
#> NA's :4962 NA's :6743ggplot(nlsy) +
geom_histogram(aes(age_bir))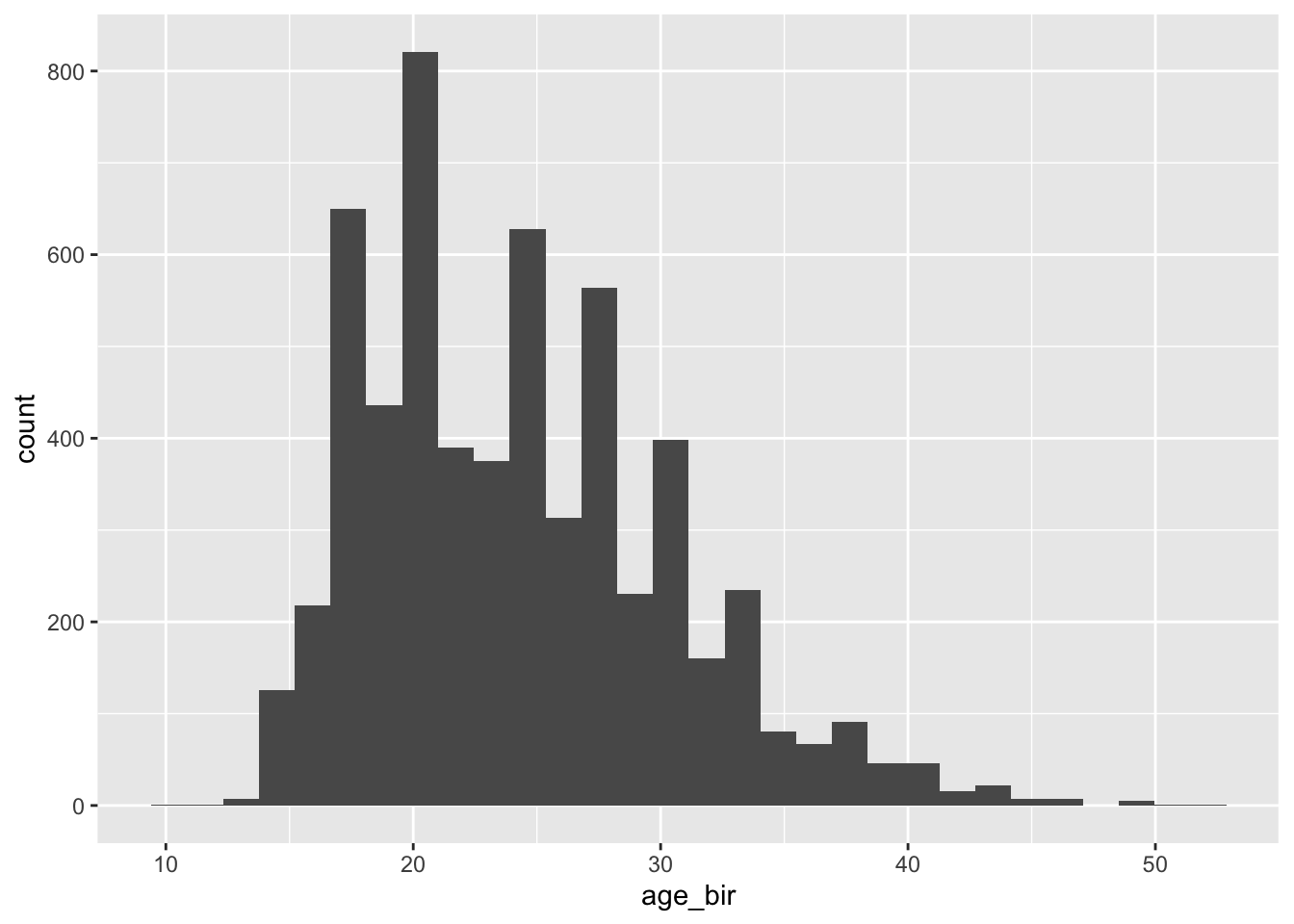
count(nlsy, nsibs)#> # A tibble: 23 x 2
#> nsibs n
#> <dbl> <int>
#> 1 0 362
#> 2 1 1611
#> 3 2 2530
#> 4 3 2412
#> 5 4 1783
#> 6 5 1198
#> 7 6 933
#> 8 7 634
#> 9 8 421
#> 10 9 289
#> # … with 13 more rowsThere are certainly some outliers we might want to investigate, but nothing else coded as missing.
(Obviously, these missing values are noted in the data dictionary. You shouldn’t have to guess what the missing values are when you’re working with real data – ask someone who knows, instead!)
Now imagine you’re interested in how the number of siblings relates to one’s age when they have their first child. Create a dataset to study this question:
- Assume that if the number of siblings is missing, they have 0
- Create a variable that is 1 if someone has kids, and 0 otherwise
- Create a dataset containing id, the sibling/child variables of interest, and income.
- Subset the data to exclude people who are missing income.
for_analysis <- nlsy %>%
mutate(
nsibs = case_when( # or ifelse(is.na(nsibs), 0, nsibs)
is.na(nsibs) ~ 0,
TRUE ~ nsibs
),
has_kids = case_when( # or ifelse(!is.na(age_bir), 1, 0)
!is.na(age_bir) ~ 1,
TRUE ~ 0
)
) %>%
select(id, nsibs, has_kids, age_bir, income) %>%
filter(!is.na(income))